The power of multi-omics
How using both transcriptomics and proteomics lets you see the bigger picture
When searching for answers to your biological questions, the technology you use is crucial. A multitude of different omics methods – including proteomics, transcriptomics, genomics, metabolomics, lipidomics and epigenomics – can be used to obtain a vast number of molecular measurements within a tissue or cell. But which omics approach will best empower your research? Here, we discuss the union of transcriptomics and proteomics and how using both of these methods can accelerate your studies.
Transcriptomics refers to the study of gene expression at the level of the transcript, which is the gene readout that occurs when DNA is copied into RNA. Analyzing the transcriptome can tell us more about the functional elements of the genome, providing information about how genes are regulated, expanding understanding of disease and revealing the molecular constituents of cells and tissues. Transcriptomics has become a popular method because of its bulk approach, growing number of methodological advancements and cost-effectiveness, allowing for experiments comparing thousands of transcripts.
However, there are drawbacks: Bulk approaches give us only a broad overview, with lower resolution, poorer sequencing depth and a lack of spatial information. Importantly, RNA does not equal protein: Proteins are created when information in DNA is transferred to mRNA, which can eventually be translated into a protein molecule. The relationship between mRNA and protein is complex and influenced by various regulatory mechanisms, and mRNA is an imperfect indicator of protein activity within the cell. Therefore, to gain true biological insights, applying transcriptomics in tandem with proteomics is key.
Proteomics is the study of the proteome – the complete set of proteins expressed by an organism – looking at the interactions, composition and structures of proteins and their cellular activities to provide a clearer understanding of an organism’s nature and function. Proteomic technologies investigate when and where proteins are expressed, how they are modified, their movements and how they interact with one another, and can shed light on which proteins interact with other proteins of interest, which proteins are involved with specific biological processes and which proteins are in particular subcellular compartments. The limitations of proteomics include difficulty in detecting low-abundance proteins and post-translational modifications, a lack of standardization in sample processing and complex analysis. Using both transcriptomic and proteomic analyses results in better, more reliable insight into the complex regulatory network of gene expression and gives you a bigger and better picture of what’s going on with your data.
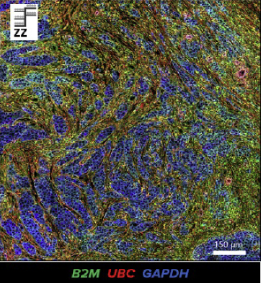
Standard BioTools™ high-throughput analysis systems enable proteomic studies to help you reveal new insights in health and disease. CyTOF® technology, which uses stable metal-tagged antibodies to convert cell counts into discrete signals, combines principles of mass spectrometry and flow cytometry to enable single-cell analysis of protein expression. Imaging Mass Cytometry™ (IMC™) uses CyTOF technology for simultaneous spatial assessment of 40-plus protein markers at subcellular resolution without spectral overlap or background autofluorescence, allowing you to image any tissue type.
In addition, a new collaboration between Standard BioTools and Advanced Cell Diagnostics builds a bridge between RNA and protein expression, which has the potential to massively increase information content of patient samples and create a new level of tissue analysis. A cutting-edge workflow combines RNAscope™ ISH technology with IMC to allow for the co-detection of RNA and protein markers in the same formalin-fixed, paraffin-embedded samples, enabling researchers to gain deeper insights into the complex cellular interactions within the tumor microenvironment (TME) and offering an opportunity to further investigations by combining transcriptomic and proteomic approaches. Applying IMC and RNAscope for spatial profiling within the TME demonstrates the value of multimodal spatial profiling, and this strategy could be applied across other fields of research for deeper insights.
Stay tuned for the next installment of this series on measuring proteomic and transcriptomic data to provide an exploration and functional analysis of the human central nervous system during glioblastoma development.
Check out other blog posts
Thanks for a wonderful 2023
Standard BioTools closed out the year with a full fall conference season. Here are some of the exciting regional events at which we had the opportunity to showcase our newest tools and technology.
A look into the pathogenic world
Equip yourself with the best tools to diagnose disease and control the course of outbreaks.
Unless explicitly and expressly stated otherwise, all products are provided for Research Use Only, not for use in diagnostic procedures. Find more information here.